MPI-LiFE
Availability of high-resolution neuroimaging and massively parallel computation support in modern HPC clusters have fueled an interest in Computational Neuroscience. This has triggered the development of algorithms to process neuroimaging data. However, most of these algorithms have been limited to a single core. In order to scale up and scale out these algorithms and bring HPC capabilities to the Neuroscience arena, we propose, MPI-LiFE; a scalable and distributed tool for statistical evaluation of brain connectomes.
MPI-LiFE is a parallel implementation of the LiFE method in the Encode Toolbox using the MVAPICH2 MPI library.
The parallel implementation uses an MPI-based distributed algorithm for sparse multiway matrix multiplication which is heavily used in the optimization algorithm in LiFE. MPI-LiFE takes advantage of optimized collectives in MVAPICH2 to drastically reduce communication cost.
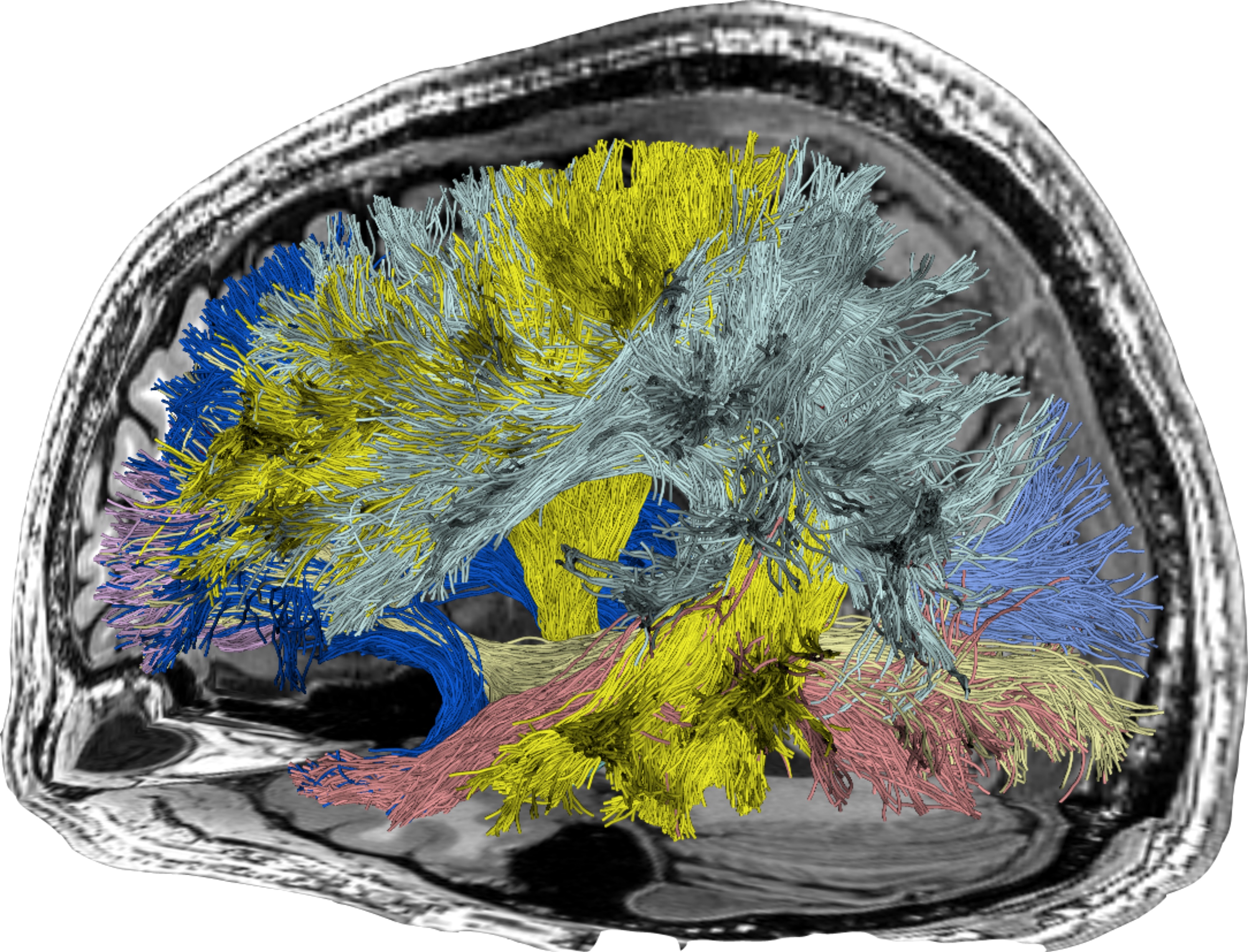
The Brain Connectome. Illustration of a set of fascicles (white matter bundles) obtained by using a tractography algorithm. Fascicles are grouped together conforming white matter tracts (shown with different colors here) connecting different cortical areas of the human brain.


